Understanding the genetic bases of human disease

Notte dei Ricercatori (Researcher’s night), 2014
People
-
-
Prof. Paola Campomenosi, e-mail to paola.campomenosi@uninsubria.it
-
-
-
Dr.ssa Raffaella Cinquetti, e-mail to raffaella.cinquetti@uninsubria.it
-
-
Dr.ssa Sarah Grossi, e-mail to sarah.grossi@uninsubria.it
We are a small, very collaborative group: from collaboration we can get the best ideas and complement expertises.
Our research is devoted to understanding the role of sequence variation or of changes in gene regulation in human disease. We study the role of gene products and the mechanisms underlying their malfunctioning in specific diseases. This in turn can lead to their exploitation in diagnosis, prognosis or in finding cures.
Here is a description of the ongoing projects, but keep in touch… We are planning more!
1. Characterization of Proline dehydrogenase, an
enzyme involved in tumorigenesis and schizophrenia.
Study of the regulation of the PRODH gene during cellular stress and identification of the factors involved in this type of regulation can help to understand the biological function of this enzyme, and how it is involved in the decision of cell fate between survival and apoptosis. Collaboration with Prof. E. Monti and M. Gariboldi (DiSTA) ; Prof. L. Pollegioni and S. Sacchi (DBSV). |
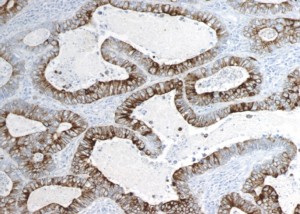
We mapped PRODH staining in human normal tissues or tumours by immunohistochemistry. We are now characterizing its expression in lung cancer (NSCLC). Our preliminary data suggest its possible exploitation for differential diagnosis of adenocarcinoma and squamocellular carcinoma. Collaboration with Prof. Fausto Sessa and Anna Maria Chiaravalli (DSCM, Anatomopathology). |
2. Quantification of circulating microRNAs as biomarkers for early diagnosis of lung cancer.
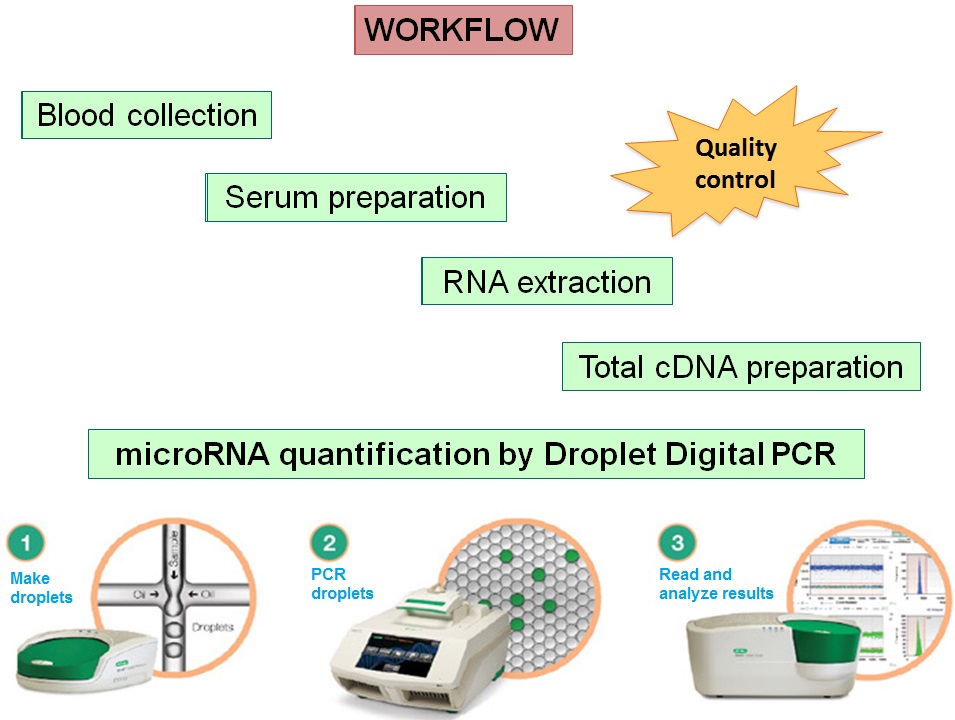
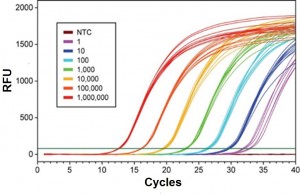
Lung cancer is one of the most common cancers, leading to death primarily because its diagnosis occurs late, when tumours are at advanced stages. Therefore, this type of tumour would greatly benefit from an early diagnosis. MicroRNAs are small RNAs, regulating gene expression, that have been shown to circulate in the blood and whose levels are altered in several human diseases. We are conducting a pilot case-control study, quantifying specific microRNAs in sera of stage I or II NSCLC patients or matched controls by Digital PCR.
Collaboration with Prof. Lorenzo Dominioni (DSCM, Università dell’Insubria), Douglas Noonan (DBSV, Università dell’Insubria) and Dr. Elisabetta Gini (PhD student); Prof. Albino Poli (Università di Verona).
3. Expression of cytokines and matrix degrading enzymes by cells grown on artificial or biological materials for abdominal wall repair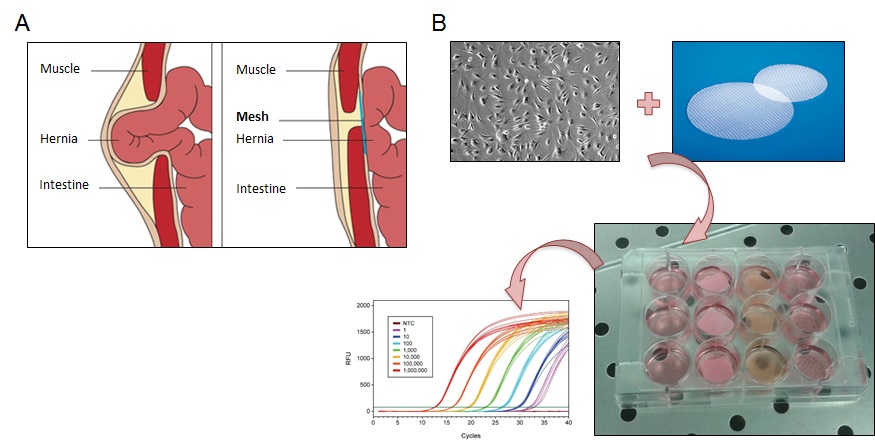
Meshes are materials used to reinforce the abdominal wall during surgery for hernia repair. They can be either synthetic or derived from decellularized biological material from different sources (e.g. porcine derma or intestinal submucosa). Their interaction with the host will determine the success of the implant. This project aims at characterizing morphology and expression of cytokines, matrix degrading enzymes and myofibroblast markers in vitro by the main cell types physiologically present at the site of implant, namely macrophages and fibroblasts. We are characterizing gene and protein expression after growth of these cells on the different materials.
Collaboration with Prof. Giampiero Campanelli (DSCM, University of Insubria), Annalisa Grimaldi (DBSV, University of Insubria).
Papers
- Moretti F., D’Antona P., Finardi E., Barbetta M, Dominioni L., Poli A., Gini E., Noonan D.M., ImperatoriA., Rotolo N., Cattoni M. Campomenosi P. 2017. Systematic review and critique of circulating miRNAs as biomarkers of stage I-II non-small cell lung cancer. Oncotarget. https://doi.org/10.18632/oncotarget.21739
- Guarino MP, Altomare A, Barera S, Locato V, Cocca S, Franchin C, Arrigoni G, Vannini C, Grossi S, Campomenosi P, Pasqualetti V, Bracale M, Alloni R, De Gara L, Cicala M. 2017. Effect of Inulin on Proteome Changes Induced by Pathogenic Lipopolysaccharide in Human Colon. PLoS One. Jan 9;12(1):e0169481.
- Guarino MPL, Vannini C, Altomare A, Barera S, Locato V, Cocca S, Arrigoni G, Alloni R, De Gara L, Campomenosi P, Cicala M. 2016. Tu1851 Protective Effect of Inulin on LPS-Induced Intestinal Smooth Muscle Impairment: A Proteomic Approach. Gastroenterology, 150(4):S960. http://dx.doi.org/10.1016/S0016-5085(16)3
- Campomenosi P, Gini E, Noonan DM, Poli A, D’Antona P, Rotolo N, Dominioni L, Imperatori A. 2016. A comparison between quantitative PCR and droplet digital PCR technologies for circulating microRNA quantification in human lung cancer. BMC Biotechnol, 16(1):60. doi: 10.1186/s12896-016-0292-7.
- Gallo S, Grossi S, Montrasio G, Binelli G, Cinquetti R, Simmen D, Castelnuovo P, Campomenosi P. 2016. TAS2R38 taste receptor gene and chronic rhinosinusitis: new data from an Italian population. BMC Med Genet. 2016 Aug 11;17(1):54. doi:10.1186/s12881-016-0321-3.
- Vannini C., Bracale M., Crinelli R., Mastrogiacomo A.R.,Campomenosi P., Marsoni M., Scoccianti V. (2014).Proteomic analysis of MG132-treated germinating pollen reveals expression signatures associated with proteasome inhibition. PLoS One, 9(9): e108811.
- Monti P., Ciribilli Y., Bisio A., Foggetti G., Raimondi I., Campomenosi P., Menichini P., Fronza G., Inga A. (2014). ΔN-P63α and TA-P63α exhibit intrinsic differences in transactivation specificities that depend on distinct features of DNA target sites. Oncotarget, 5, 2116-2130.
- Cappelletti P., Campomenosi P., Pollegioni L. and Sacchi S. (2014). On the degradation of human D-amino acid oxidase and its interacting partner pLG72, two key proteins in D-serine catabolism in the brain. FEBS Journal, 281, 708-723.
- Raimondi I., Ciribilli Y., Monti P., Bisio A., De Sanctis V., Pollegioni L., Fronza G., Inga A. and Campomenosi P. (2013). P53 family members modulate the expression of PRODH, but not PRODH2, via intronic p53 response elements, PLoS One, 8(7): e69152.
- Campomenosi P., Cinquetti R., Tallarita E., Lindqvist C., Raimondi I., Grassi P., Näsman J., Dell A., Haslam S.M., Taramelli R. and Acquati F. (2011). Comparison of the baculovirus insect cell and Pichia pastoris heterologous systems for the expression of the human tumor suppressor protein RNASET2. Biotechnology and Applied Biochemistry, 58: 39–49.
- Acquati F., Bertilaccio S., Grimaldi A., Monti L., Cinquetti R., Bonetti P., Lualdi M., Vidalino L., Fabbri M., Sacco M.G., Rooijen N., Campomenosi P., Vigetti D., Passi A., Riva C., Capella C., Sanvito F., Doglioni C., Gribaldo L., Macchi P., Sica A., Noonan D.M., Ghia P. and Taramelli R. (2011). Microenvironmental control of malignancy exerted by RNASET2, a widely conserved extracellular RNase. Proc Natl Acad Sci U. S. A. 108, 1104-1109.